|
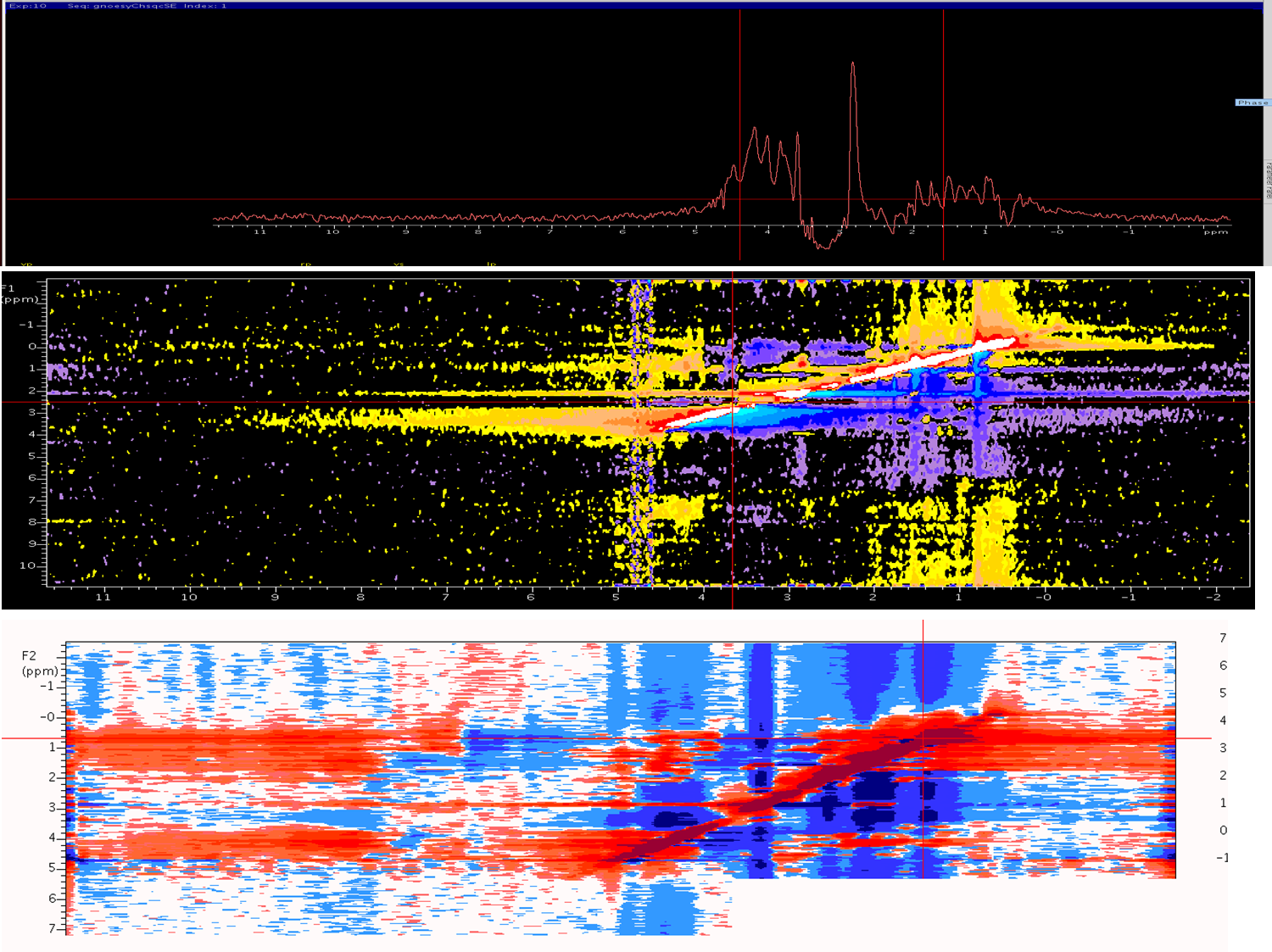
Dear NMR Wiki'ers I Have acquired c13-edit noesy experiment on 700vnmr for my target protein (20kd) with mixing time of 120ms , I tried my best to correct the phasing, still it seems to be negative contours ,Peak broadening and noise is more after processing . How to overcome this problem ? Fig-1 1D projections fig-2 f1f3 projection in process in t1 fig-3 f1f3 projection in process in t1 after transformation (1026-512-256)
|
|
I know a commercial platform that provides custom protein services, such as crystallization, structure determination and function analysis by X-ray, EM, NMR, etc. The platform provides MagHelix™ STD NMR technique for characterization of the binding of a ligand to its receptor protein. STD NMR is a powerful and fast method used to study protein-ligand interactions, focusing on the signals of the ligand. Besides, values including KD, the association and dissociation kinetics (Koff and Kon respectively) can be obtained from STD NMR study as well. |
|
If f1180='y', the first time point in the indirect dimension starts at one-half the dwell time rather than zero, and the phase should be rp1=90 and lp1=-180. In NMRpipe the phases should be -90 and 180. You may still need to slightly touch up the phasing as described by George. |
|
You don't say what spectrometer or processing software you are using. However, in general if this is a 3D data set you can process the first full F1F3 2D plane (in VnmrJ use wft2da(1)). I'd first set the zero-order and first-order phases to zero (lp1,rp1). Then you can phase the data by selecting a horizontal slice at the upper right and displaying it (use the F1F3 page in the Display folder-use the "Show 1D Slice" button at upper right in the panel). Then phase the spectrum using the mouse in the normal way, but only to change rp1. Then redisplay the 2D F1F3 data and select a slice for a peak in the lower left. Again, select the "Show 1D Slice", click at the right and click at the left. Phase up the spectrum peaks at the left (this sets lp1). You may have to repeat the rp1/lp1 phasing steps until the spectrum is phased. You should be able to phase the data in F3 by doing wft(1) and phasing normally. It looks just like your phasing is off, giving you negative portions of your lines George Gray Agilent Technologies I am using BioPack/VNMRJ. - sri (Nov 08 '11 at 01:49) |