|
Hi, we've tried to apply SOL function of nmrPipe to process a 3D 13C edited NOESY. and the result is a wide antiphase-looking peak on water resonance that strongly distorts the baseline. Would you have any idea what might be causing this? First image is a screenshot of data using SOL processing step and the second one was prepared without SOL. Thanks.
|
|
Based on the documentation of NMRpipe it appears that the SOL function calculates the average over the chosen window and subtracts it out from the current datapoint. The length of the window can have a dramatic effect on the quality of the solvent suppression. Based on this, and a quick look at the NMRpipe documentation, I would expect that a substantial improvement of solvent suppression can be obtained by using a lower the value of flenpts parameter associated with the SOL function. In addition, I would expect that changing the fshape parameter to select for sinusoidal weighting (instead of the default boxcar) would be helpful. The mirror option of the SOL function may also be useful. My interpretation of your artefact is as follows (though I have not seen the code for NMRpipe): the algorithm calculates the (weighted) average over the chosen window and subtracts it out from the current fid. The problem is that, this is not possible for the first few data-points; the window terminates at zero time, and the data for calculating the average is not available for the first few points. Therefore, the subtracted fid contains a solvent artifact contributed from the first few data points. There are several ways to overcome this problem:
Note: I have never used NMRpipe (I used Felix and data processing software written by myself). |




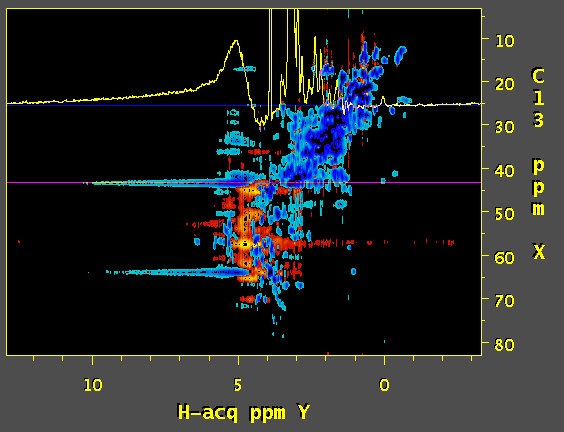
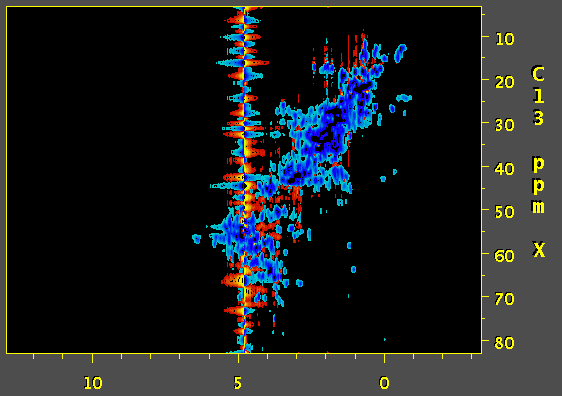

Have you made sure that the offset is perfectly on the water resonance. This is important for both the 'SOL' and 'POLY -time' function. Have a look at this: http://tech.groups.yahoo.com/group/nmrpipe/message/526 - TJCarruthers (Apr 11 '11 at 23:19)