|
Hello, I know that this problem has been discussed a number of times on the nmrPipe newsgroup and there are some helpful hints (especially here), but some details seem to be missing. In the conversion script I have The beginning of my processing script looks like this: everything else is the same as in standard hsqc processing and below is the picture that we get in nmrDraw. Sign of the 15N frequency is not discriminated. Any help will be appreciated! Thanks.
|
|
I found a post post from Frank Delaglio on the NMRPipe message board that answers the same question, but I think there was a confusion about ordering of the IP/AP loop with respect to the quadrature frequency discrimination loop. This solution worked for the setup where The usual 2D conversion script (but notice the To process the data you can use that same method that is normally used to process HSQC,
but with one line inserted right after ... first obtain one subset: ... then change the Then calculate the final separated spectra. Oh, wow. Sorry it condensed the script like--I'm not sure how to get it to preserve the spaces after the \'s when I paste. - rshapiro (Dec 11 '09 at 17:00) the editor button with ones and zeroes formats source code - Evgeny Fadeev (Dec 11 '09 at 17:20) |
|
This question might have been better posted in the NMRPipe group. Normally, in NMRPipe, gradient enhanced Varian 2D data is converted with "-yMODE Rance-Kay". This actually runs an NMRPipe macro to perform gradient shuffling of pairs of adjacent 1D vectors during the conversion, but it is run "in background", and you don't see it explicitly: In an interleaved version of the gradient-enhanced data, there are two possible formats. In the case array="phase,IPAP", the innermost acquisition loop is the Rance-Kay phase encoding. So, adjacent pairs of vectors are Rance-Kay pairs, and you can convert this data with "-yMODE Rance-Kay". Then, later processing steps can use COADD to select or combine channels. However, when array='IPAP,phase' the IP/AP alternation is in the innermost acquisition loop. This means that manipulation with COADD must happen before gradient shuffling. In these cases, we convert with "-yMODE Complex", and then explicitly include gradient shuffling just afterwards, for example: Also, one more tip: When dealing with this kind of data, it can be handy to extract the IP and AP channels separately ("COADD -cList 1 0" and "COADD -cList 0 1") so that they can be inspected first to find the optimum coefficient for cancelation of IP and AP signals. Then, use the stand-alone program "addNMR" to combine the two channels to form scaled sum and difference, for example: Thanks, Frank. This doesn't work for my dataset unfortunately. I suspected that when you do COADD on array=IPAP,phase data you will be adding wrong vectors, i.e. just real and imaginary of the same subset (IP or AP). Tried header manipulation with REAL then coadd then back to complex, but no avail. - Evgeny Fadeev (Dec 10 '09 at 07:25) we did in the end sort this out similarly, but with using "| nmrPipe -fn COADD -cList 1 -1 -time" after "nmrpipe -fn TP". No idea why this makes a difference. Also gNhsqc_IPAP does not use Rance-Kay shuffling. - Evgeny Fadeev (Dec 11 '09 at 09:36) also, macro var_ranceY.M doesn't seem to be applicable for this particular sequence. - Evgeny Fadeev (Mar 03 '10 at 09:58) |




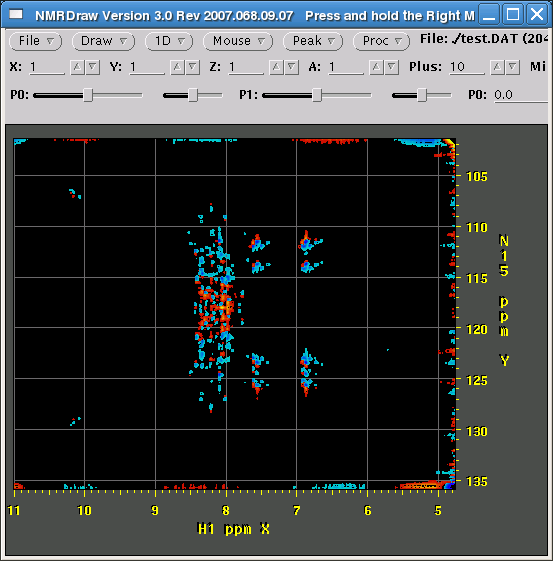

should I have had array='phase,IPAP' instead? If so is there a way to recover here using NMRPipe? Thanks! - Evgeny Fadeev (Dec 06 '09 at 10:21)