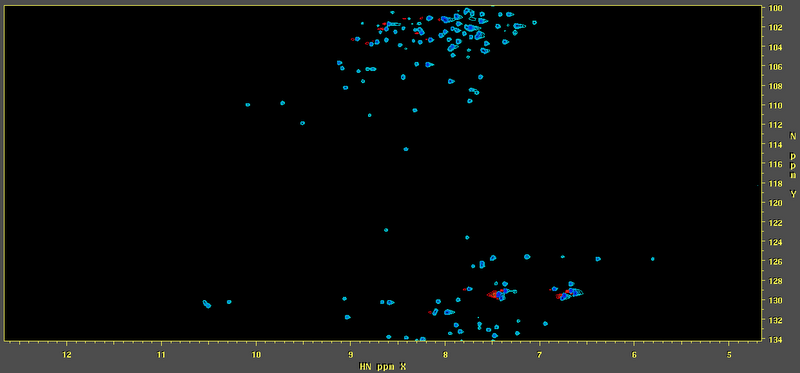
i'm trying to process an HSQC by using standard scripts of NMRpipe, and get this:
https://picasaweb.google.com/Nadia.K.W/NMR#5593296316808640578
what is wrong in my processing? at the spectrometer the spectrum looked nice, so it's a matter of processing...
conversion:
bruk2pipe -in ../Bruker/ser -bad 0.0 -aswap -DMX -decim 2080 -dspfvs 20 -grpdly 67.9842 \ -xN 4096 -yN 512 \ -xT 2048 -yT 256 \ -xMODE DQD -yMODE States-TPPI \ -xSW 9615.38 -ySW 2100 \ -xOBS 599.762823 -yOBS 60.780 \ -xCAR 4.707 -yCAR 115.00 \ -xLAB HN -yLAB N \ -ndim 2 -aq2D States-TPPI \ -out ../Data/test.fid -verb -ov
processing:
nmrPipe -in ./test.fid \ | nmrPipe -fn POLY -time \ | nmrPipe -fn SINE -c 0.5 -off 0.45 -end 0.98 -pow 2 \ | nmrPipe -fn ZF -auto \ | nmrPipe -fn FT \ | nmrPipe -fn PS -p0 -20.0 -p1 0.0 -di \ | nmrPipe -fn EXT -left -sw \ | nmrPipe -fn TP \ | nmrPipe -fn SINE -c 0.5 -off 0.45 -end 0.98 -pow 2 \ | nmrPipe -fn ZF -auto \ | nmrPipe -fn FT \ | nmrPipe -fn PS -p0 0.0 -p1 0.0 -di \ | nmrPipe -fn TP \ | nmrPipe -out ./test.ft -ov
thanks for the tips!
Nadia