Dear NMR Wiki'ers
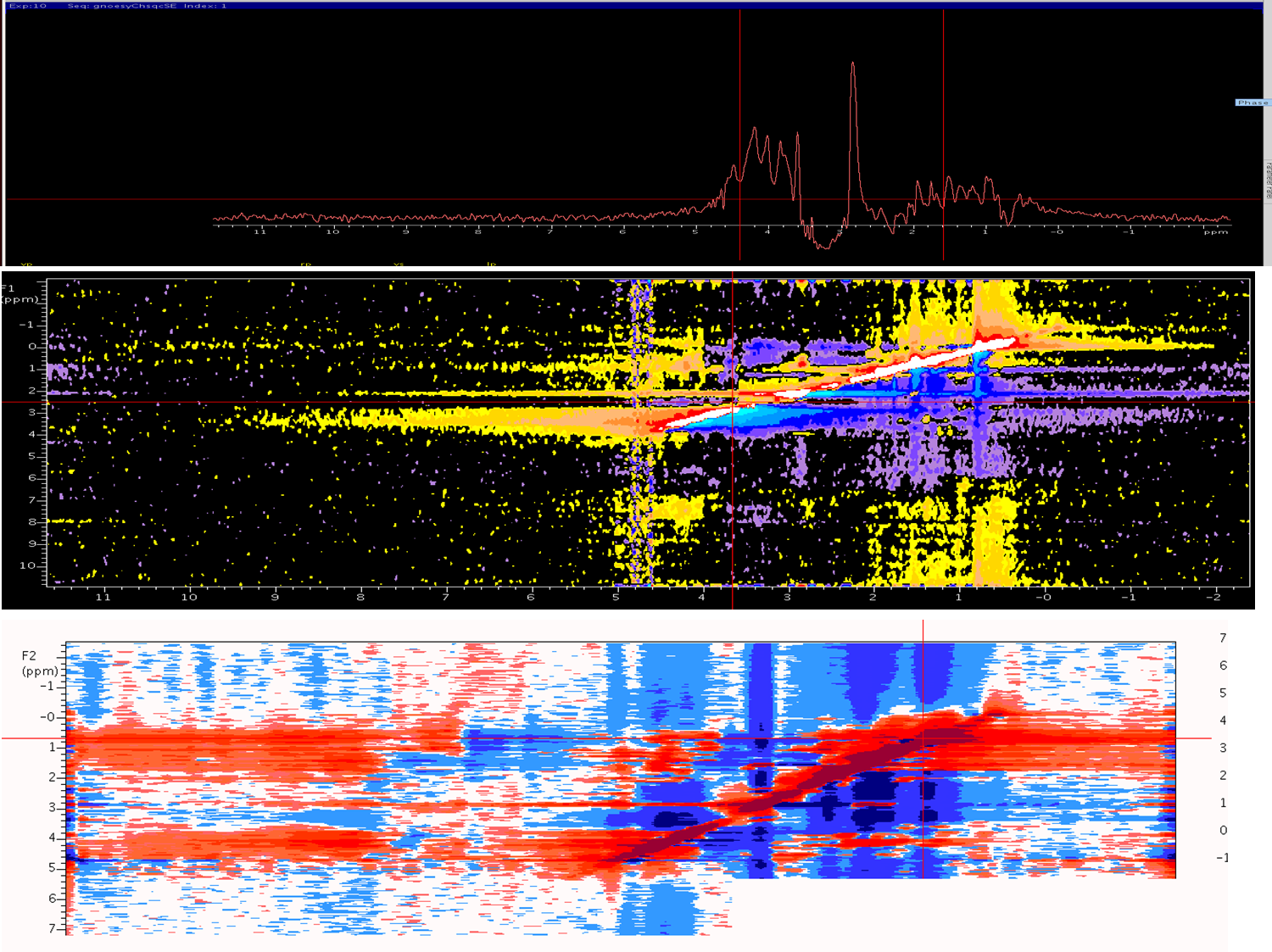
I Have acquired C13 EDIT NOESY (C13,N15,H1) experiment on 700vnmr for my target protein (20kd) with mixing time of 120ms , I tried my best to correct the phasing, still it seems to be negative contours ,Peak broadening and noise is more after processing . How to overcome this problem ?
Fig-1 1D projections fig-2 f1f3 projection in process in t1 fig-3 f1f3 projection in process in t1 after transformation (1026-512-256)